Cryo-electron tomography (cryoET) allows for observation of in situ structures of biological macromolecules within cells, and has attracted significant attention in recent years. During cryo-electron microscopy imaging, the contrast transfer function (CTF) affects the collected micrographs, which thereby prevents them from directly reflecting the original structural information. Accurately estimating defocus parameters based on the CTF is crucial for eliminating the influence of CTF, which is essential for image quality assessment and high-resolution structure analysis. Currently, commonly used defocus estimation methods typically rely on the CTF diffraction rings (Thon rings) in the power spectrum of the micrograph. However, micrographs in cryoET tilt series have very low signal-to-noise ratios (SNR). Meanwhile, due to sample tilting, the defocus changes continuously with the height of the sample. These factors result in fuzzy Thon rings in the power spectra of the micrographs taken at high tilt angles, making it difficult to accurately estimate the defocus values. Additionally, lamellar samples prepared by cryo-focused ion beam (cryoFIB) milling often have an angle of around 10 degrees between the sample and the stage, known as the absolute tilt angle offset. This offset affects the estimation of the local defocus values, thereby influencing the overall accuracy of defocus estimation.
On May 31, 2024, the research groups led by Xueming Li and Yuan Shen, respectively, jointly published a work online in Structure, titled “Tilt-series-based joint CTF estimation for cryo-electron tomography.” This work reports a joint estimation method based on all the micrographs in the entire tilt series (Figure 1). The method first utilizes micrographs in the entire tilt series to estimate the average defocus of the whole dataset. By introducing a Thon ring rescaling technique to scale the power spectra under different defocus values to a common reference, different Thon rings can be aligned and averaged, thereby obtaining a high SNR spectrum of Thon rings. Then, the method uses the average defocus as the initial value to perform local refinement of the defocus for each individual micrograph in the tilt series. During the estimation process, the method can simultaneously estimate the absolute tilt angle offset based on the grayscale mean of the micrograph and the off-plane tilt angle of the tilt axis (often known as x-tilt) jointly. Involving more variables in the estimation enhances the estimation accuracy of the defocus.
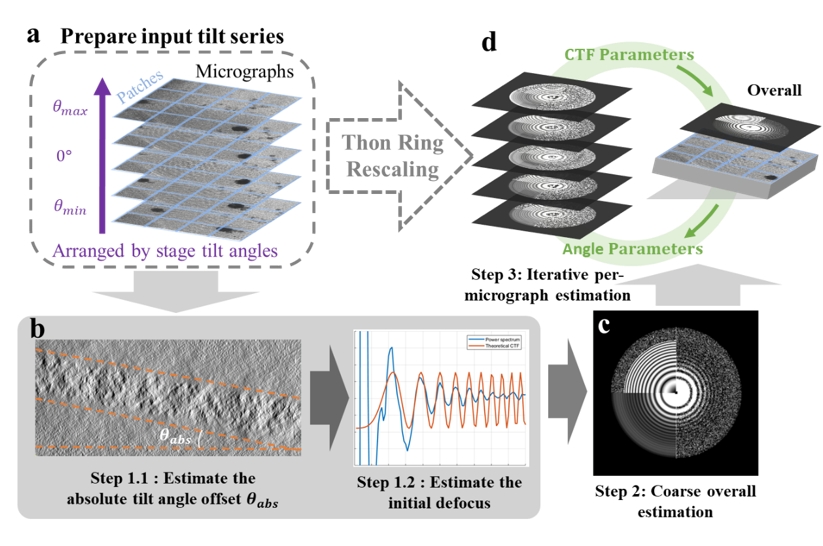
Figure 1. The workflow of CTFMeasure.
Based on the joint estimation method above, the research team developed a new software called CTFMeasure. Tests show that, compared to the existing methods, clearer Thon ring patterns in the power spectrum can be visualized by CTFMeasure (Figure 2). By leveraging this, it not only achieves accurate defocus estimation but also provides support for users to assess data quality based on the quality of the Thon rings. Experiments show that the defocus estimated by CTFMeasure can provide more accurate initial values for subtomogram averaging, which offers strong support for high-resolution structure determination. A better initial defocus value also helps reduce the number of iterations required for three-dimensional (3D) refinement of the structure, which improves computational efficiency (Figure 3).
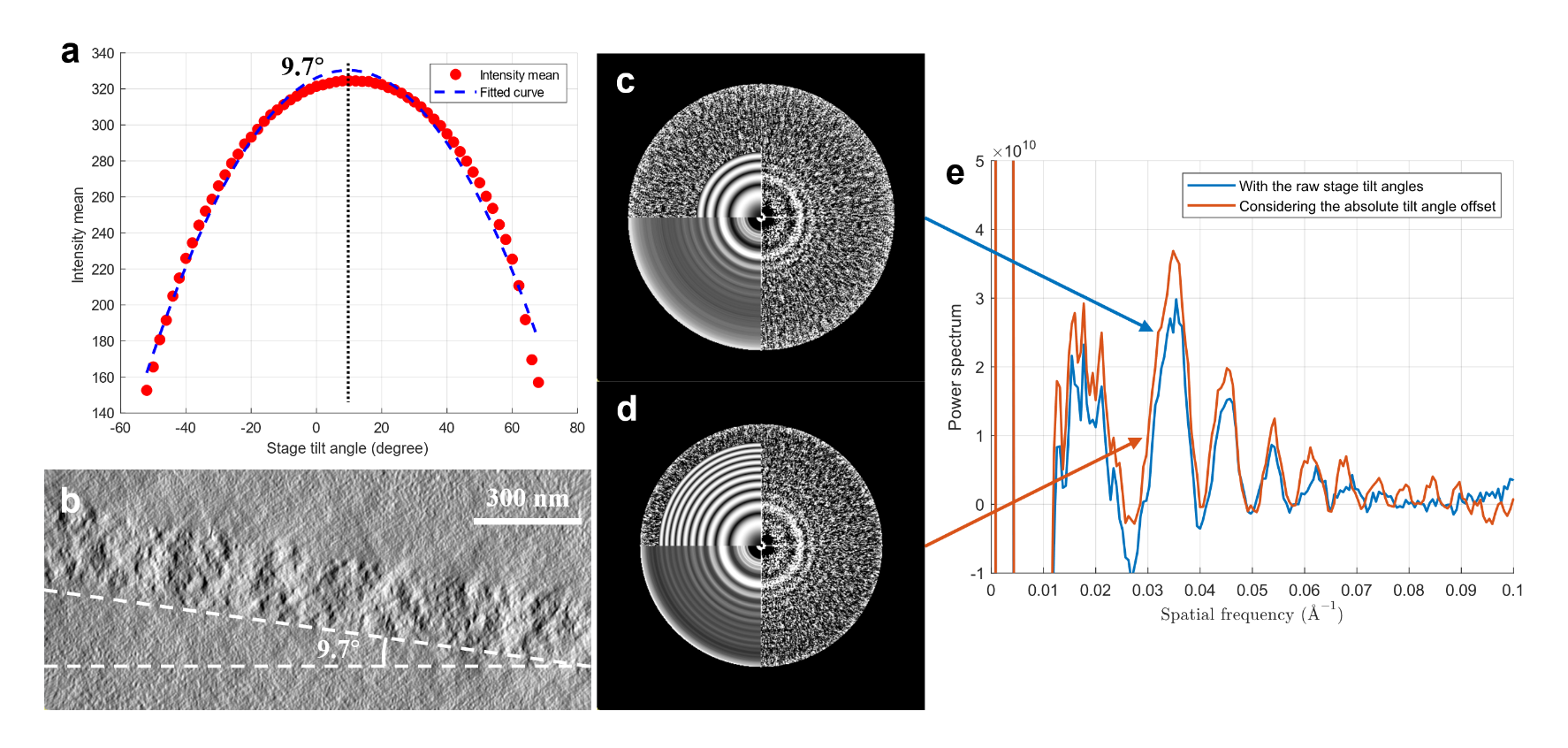
Figure 2. Absolute tilt angle offset estimation and comparison of Thon rings after rescaling.
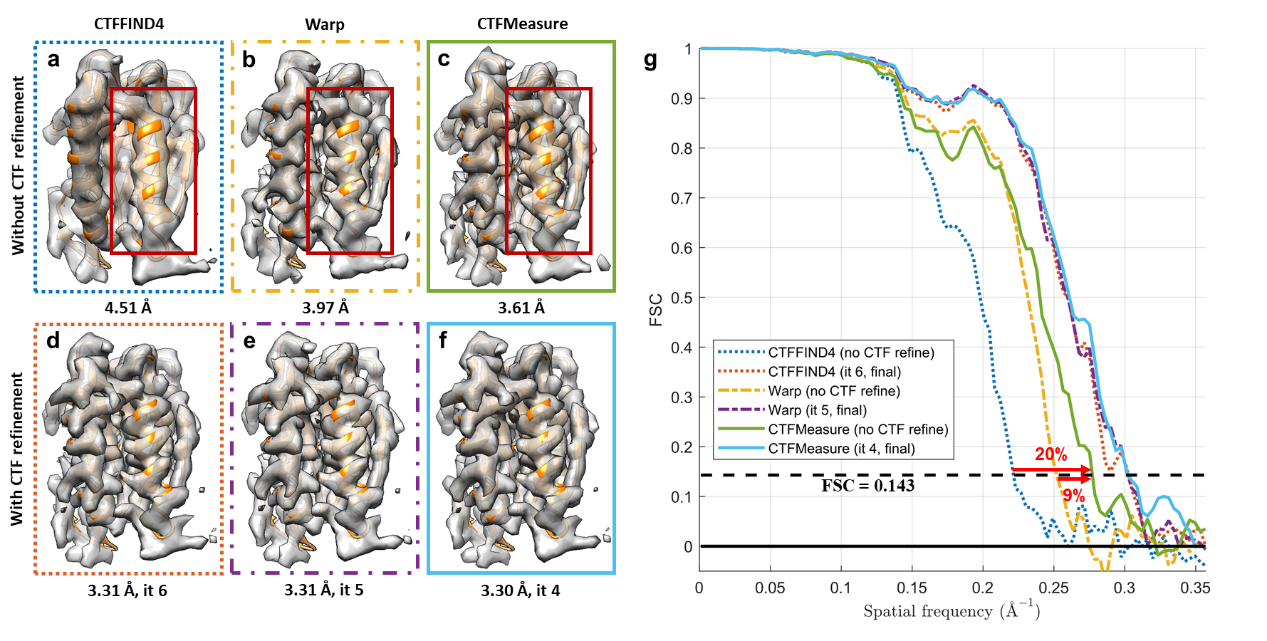
Figure 3. Comparisons of subtomogram averaging with defocus values estimated by different methods.
Associate Professor Xueming Li from the School of Life Sciences and Professor Yuan Shen from the Department of Electronic Engineering are the corresponding authors of this paper. Mr. Ranhao Zhang is the first author of the paper, who is a PhD student in the Department of Electronic Engineering, jointly supervised by Li and Shen. All the authors come from Tsinghua University. This work was funded by the National Natural Science Foundation of China, Tsinghua-Peking Joint Center for Life Sciences, and Beijing Frontier Research Center for Biological Structure. This work was also supported in computing and cryoEM instruments by the Tsinghua University Branch of China National Center for Protein Sciences Beijing.
Link to the paper:https://doi.org/10.1016/j.str.2024.05.006
Editor: Li Han

