Yigong Shi’s group reports on the structure of autosomal dominant polycystic kidney disease related to the PKD1/PKD2 complex
The kidney is an important living organ of the human body. It has many physiological functions such as excreting metabolic products, regulating water and electrolyte balance and the endocrine system. Under a variety of pathological conditions, the kidneys need to excrete blood from the organs and supply it to more important organs (e.g. the heart and brain). While of such great importance, the kidneys, however, are among those most vulnerable organs. Genetic factors, hyperglycemia and hyperlipidemia are important causes of chronic kidney disease. According to the National Institute of Health, the prevalence of chronic kidney disease in US adults (approximately 200 million in total) has reached 11.3%.
Autosomal dominant polycystic kidney disease (ADPKD) is one of the most important causes of chronic kidney disease, with an incidence of 1/400-1/1000. About 12 million patients worldwide are affected by this disease. The bilateral kidneys of the patient gradually produce fluid-filled vesicles with increasing age, squeezing and destroying the surrounding normal tissues. About 50% of patients develop end-stage renal failure, requiring heterologous kidney transplantation or life-long hemodialysis. There are about 1.5 million patients with this disease in China. Every year, tens of thousands of patients are on the waiting list for donated kidney transplantation or sustaining life through continuous dialysis. ADPKD not only causes severe physical and mental suffering to the patient, but also imposes a heavy financial burden on the patient's family.
The genes associated with ADPKD pathogenesis are PKD1 and PKD2, and their gene products are the membrane proteins PKD1 and PKD2 respectively. Mutations in both accounted for approximately 85% and 10% of all patients respectively. The human pkd1 gene is located on chromosome 16, encoding a protein of PKD1 with a length of 4302 amino acids containing 11 transmembrane helices. Because of the huge molecular weight of PKD1, researchers have been challenged with great technical difficulties in their research. Since the successful sequencing of the pkd1 gene in 1993, many scientists have been investigating this protein for more than 20 years. Although these studies have broadened the perception of ADPKD, the function of the PKD1 protein and the pathogenesis of polycystic kidney disease remains controversial for lack of sufficient information. Another pathogenic protein, PKD2, is a chaperone molecule of PKD1, and plays an extremely important role in the folding of PKD1, transporting among organelles and protein maturation. PKD1 and PKD2 proteins can interact to form hetero-tetrameric complexes and may perform important physiological functions on primary cilia.
On August 10th, 2018, UTC+8, Yigong Shi's group, published a research article entitled "Structure of the human PKD1 and PKD2 complex" online in the journal Science, reporting the first near-atomic resolution (3.6 ?) of the polycystic kidney disease-associated protein PKD1/PKD2 complex.
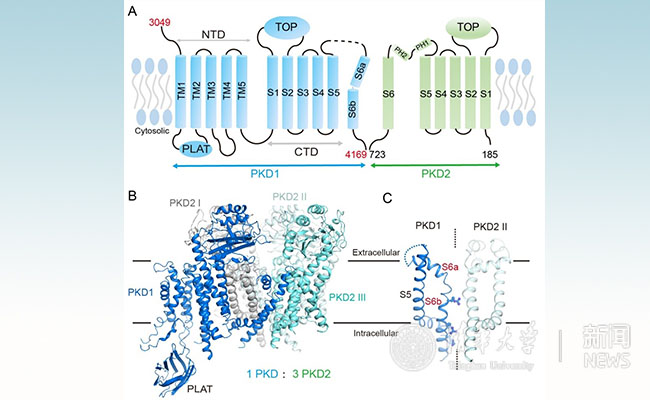
Figure 1: A. schematic diagram of the topology of human PKD1 and PKD2 proteins. B. The overall structure of human PKD1 and PKD2 protein complex; C. The unique channel domain of PKD1.
Yigong Shi's group first resolved the near-atom resolution structure of human PKD1 and PKD2 complexes. This structure reveals that the PKD1 and PKD2 proteins form a distinctive one-to-three complex (1 PKD1: 3 PKD2). Based on this structural and biochemical data, the team found that PKD1 and PKD2 were able to form complexes without the protein C-terminal coiled-coil domain. This result is inconsistent with the mainstream doctrine which previously thought that “no protein complex can be formed when there is no coiled-coil domain”. Therefore, many studies based on this conclusion need to be reconsidered. In addition, the researchers found that the pore domain structure of PKD1 is different from that of all currently known voltage-gated ion channels. The S6 transmembrane helix of PKD1 has a number of positively charged amino acids protruding into the central cavity of the channel, potentially blocking the central channel path which is for calcium permeation. There have been many debates about whether PKD1 and PKD2 form calcium channels in the field. The current conformation of this structure does not support the channel hypothesis, which brings new thinking to the investigation of the mechanism of polycystic kidney disease.
Yigong Shi's research team cooperated with Prof. Mei Changlin and Prof. Yu Shengqiang from Shanghai Changzheng Hospital. Since 2013, the structure of two human proteins, PKD1 and PKD2, has been conceived. During the past five years, unremitting efforts on the protein boundary, frozen sample preparation conditions and detergent optimization have been tried and screened. Finally, the structure of PKD1 and PKD2 protein complex was resolved with an overall resolution of 3.6 ?, and the core region resolution was able to reach 3.2 ?. This was the first time that the structure of a TRP channel family heterologous complex had been obtained.
Professor Yigong Shi from the School of Life Sciences of Tsinghua University and the Center for Structural Biology and Innovation, is the corresponding author of this article; Qiang Su, a third-year doctoral student at the School of Life Sciences of Tsinghua University, and Dr. Feizhuo Hu from the School of Medicine,, are the co-first authors of this article; Ge Xuefei, an undergraduate student in the six-character class of the School of Life, Tsinghua University, helped complete some of the experiments. Dr. Lei Jianlin from Tsinghua University's cryo-electron microscopy facility provided assistance in the collection of cryo-electron microscopy data. Professor Zhou Qiang, School of Medicine, Tsinghua University, provided cryo-EM data processing guidance. Associate Professor Wang Tingliang, School of Medicine, Tsinghua University, was involved in the early operation of the subject. The electron microscope data was collected from the cryo-electron microscope facility of Tsinghua University. The calculation work was supported by the Tsinghua University High Performance Computing Platform and National Protein Facility Experimental Technology Center (Beijing). This work has received funding support from the Beijing Center for Structural Biology and the National Natural Science Foundation.
Research article link: http://science.sciencemag.org/content/early/2018/08/08/science.aat9819
Contributor: School of Life Sciences
Editors: John Olbrich, Guo Lili

