Yigong Shi’s Group Elucidated the cryo-EM structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation
On May 25th 2018, Professor Yigong Shi’s group in the School of Life Sciences, Tsinghua University, published an article entitled “Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation” in the prestigious international magazine Science. The paper reported the cryo-EM structures of the precursor pre-catalytic spliceosome (the pre-B complex) and pre-catalytic spliceosome (the B complex) in Saccharomyces cerevisiae, at the average resolutions of 3.3-4.6 angstrom and 3.9 angstrom, respectively. These structures of the two crucial complexes for RNA splicing before activation represent an important step towards a complete dissection of the spliceosome assembly, activation, and catalysis.

In 1977, scientists observed that adenovirus mRNA could not form a continuous double-stranded RNA-DNA hybrid duplex with its corresponding DNA transcription template. Instead, single-stranded DNA bulges were extended at different positions in the hybridized double-stranded RNA-DNA duplex. This discovery suggested that the transfer of genetic information from DNA to mRNA not only involved transcription but also pre-mRNA splicing, which is to further remove the non-coding regions (introns) and connect the coding sequences (exons). RNA splicing is ubiquitous in eukaryotes. The average numbers of introns per gene increase from simple single-cell eukaryotes, such as yeast, to higher organisms such as worms and flies, and all the way up to humans. Some pre-mRNAs can be spliced in more than one way. Therefore, mRNAs containing different combinations of exons can be generated from a given pre-mRNA.
Pre-messenger RNA (pre-mRNA) splicing is a crucial and the most complex step in the central dogma in eukaryotic cells. This process is executed by a dynamic mega-complex known as the spliceosome. From the discovery of RNA splicing in 1977 to the beginning of this century, scientists have recapitulated assembly and disassembly of the spliceosome by immunoprecipitation, gene knockout, cross-linked mass spectrometry, and in vitro splicing reaction and so on. The essence of RNA splicing is the removal of intron and connection of exons through two successive transesterification reactions in which phosphodiester linkages within the pre-mRNA are broken and a new one is formed. To catalyze splicing of the pre-mRNA, spliceosome has to undergo extremely precise stepwise assembly by forming a series of spliceosomal complexes. Based on the assembly and catalytic states and the biochemical composition of the spliceosome, these spliceosomal complexes have been defined as the A, pre-B, B, Bact , B*, C, C*, P, and ILS complexes.
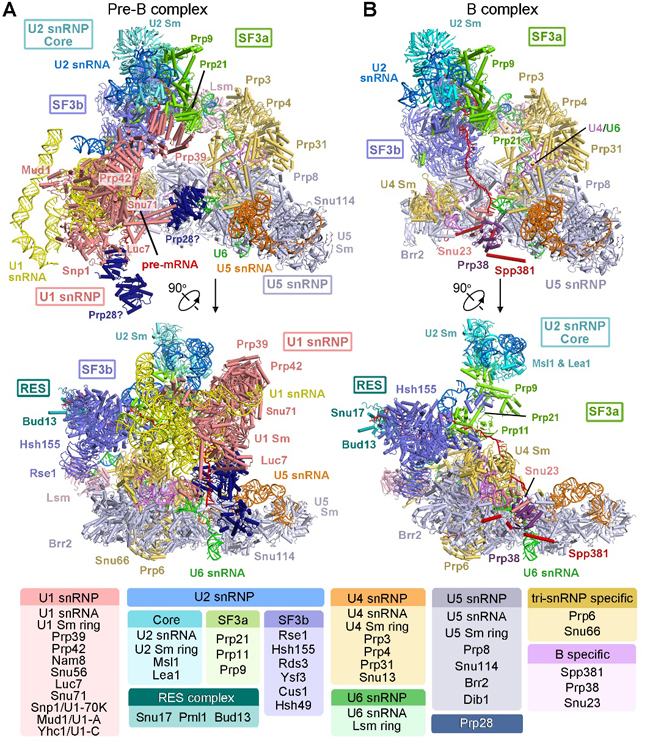
Figure 1. Structures of precursor pre-catalytic spliceosome (pre-B complex) and pre-catalytic spliceosome (B complex) from S. cerevisiae.
Precursor pre-catalytic spliceosome (pre-B complex) is a highly dynamic and transient complex that is difficult to obtain under normal physical conditions. In their recent Science paper, Shi and colleagues enriched pre-B complex samples from S.cerevisiae cells through over-expression of an ATPase-defective Prp28 mutant, leading to the blockage of the dissociation of U1 snRNP in vivo. Then they acquired the good pre-B and B complexes by applying TAP purification strategy and determined the three-dimensional structures by single particle cryo-EM, and built the atomic model (Figure 1). Structure of the S. cerevisiae pre-B complex contains 68 discrete proteins, five snRNA molecules, and the pre-mRNA. Structural elucidation of the pre-B complex fills an important void in the mechanistic understanding of pre-mRNA splicing by the spliceosome. The local resolutions of the core of U1 snRNP and the tri-snRNP reach 3.0 angstrom, which enabled assignment of atomic features. Advent of the pre-B structure, along with other published information, reveal the molecular mechanism for assembly and activation of the S. cerevisiae spliceosome. Remarkably, the structure of pre-B complex for the first time revealed the recognition mechanism of 5’SS of the pre-mRNA by U1 snRNP. The authors propose that the structure of the A complex may be faithfully represented in the pre-B complex (Figure 2). Therefore, these two structures provide important structural evidence to illustrate the mechanism of 5’SS recognition by U1 snRNP and the spliceosome assembly and activation, which collectively advance our understanding of pre-mRNA splicing.
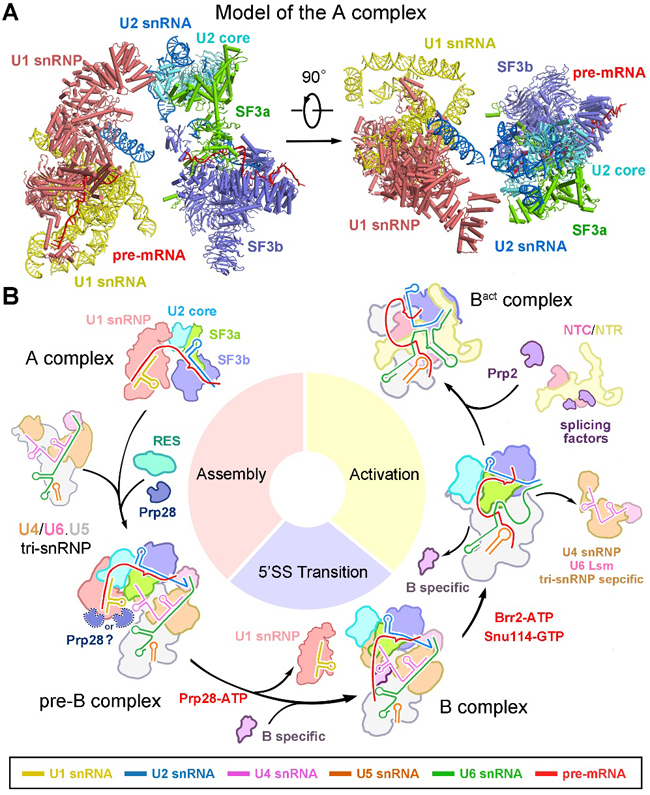
Figure 2. Mechanism of assembly and activation of the spliceosome in S. cerevisiae.
The Shi Group has been focusing on the structural and mechanistic elucidation of pre-mRNA splicing in the past decades. After the success of crystal structural elucidation of subcomplexes within spliceosome, they took a direct assault on the intact spliceosome and made the breakthrough in the summer of 2015. Since then, the Shi lab has reported nine high resolution structures of the crucial spliceosomal complexes, including the S.pombe intron lariat spliceosome ILS complex at 3.6 angstrom, the pre-assembled U4/U6.U5 tri-snRNP at 3.8 angstrom, the activated spliceosome Bact complex at 3.5 angstrom, the catalytic step I spliceosome C complex at 3.4 angstrom, the step II catalytically activated spliceosome C* complex at 4.0 angstrom, the post-catalytic spliceosomal P complex at 3.6 angstrom, the ILS complex at 3.5 angstrom from S.cerevisiae and the newest published precursor pre-catalytic spliceosome (pre-B complex) and pre-catalytic spliceosome (B complex) at 3.3-4.6 and 3.9 angstrom, respectively. These nine high resolution structures nearly cover all the essential functional states for spliceosome assembly, catalysis and disassembly, which provide unprecedented insight into the chemical basis for RNA splicing (Figure 3).
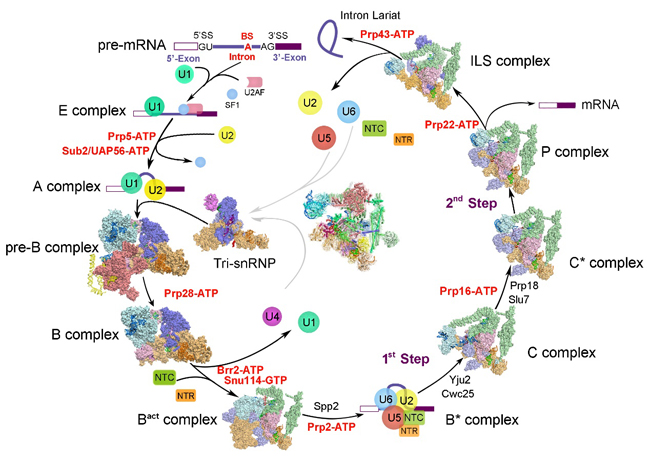
Figure 3. Cryo-EM structure of yeast spliceosome solved by Shi lab.
Prof. Yigong Shi is the corresponding author. Rui Bai (3th-year PhD student from the School of Life Sciences), Dr. Ruixue Wan (post-doc fellow from the School of Medicine), and Dr. Chuangye Yan (post-doc fellow from the School of Life Sciences and Center for Life Sciences) are the co-first authors of this paper. Dr Jianlin Lei provided technical support on the EM data collection. EM images were acquired at the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing). Data processing was performed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology, the Computing Platform of China National Center for Protein Sciences (Beijing). This research was funded by the Beijing Innovation Center for Structural Biology and the National Natural Science Foundation of China.
The original link:
http://science.sciencemag.org/content/early/2018/05/23/science.aau0325
Related publications:
http://science.sciencemag.org/content/early/2016/01/06/science.aad6466
http://science.sciencemag.org/content/early/2015/08/19/science.aac8159
http://science.sciencemag.org/content/early/2015/08/19/science.aac7629
http://science.sciencemag.org/content/early/2016/07/20/science.aag0291
http://science.sciencemag.org/content/early/2016/07/20/science.aag2235
http://science.sciencemag.org/content/early/2016/12/14/science.aak9979.full
http://www.cell.com/cell/fulltext/S0092-8674(17)30954-6
http://www.cell.com/cell/fulltext/S0092-8674(17)31264-3

