Yigong Shi’s group reported the first atomic structure of human spliceosome in a Cell research article
A research team led by Professor Yigong Shi from the School of Life Sciences at Tsinghua University has published a research article in the journal Cell, reporting the first atomic structure of the human spliceosome at 3.8 angstrom resolution that was determined by single particle cryo-electron microscopy. This structure represents the conformation of the spliceosome just prior to exon ligation and reveals important insights into step 2 transesterification reaction.
In eukaryotes genes are not continuous. Instead they are interrupted by non-coding sequences, known as introns that have to be removed during gene expression. Splicing of the pre-mRNA is executed by an enormous and highly dynamic macromolecular machine called spliceosome. Spliceosomal remodeling by the conserved DExD/H-box ATPases/helicases during each splicing cycle generates at least seven distinct states: the pre-catalytic B complex, the activated Bact complex, the step I catalytically activated B* complex, the C complex, the step II catalytically activated C* complex, the post- catalytic P complex, and the ILS complexes (Figure 1). Defects in pre-mRNA splicing can impair gene expression and lead to human genetic diseases.
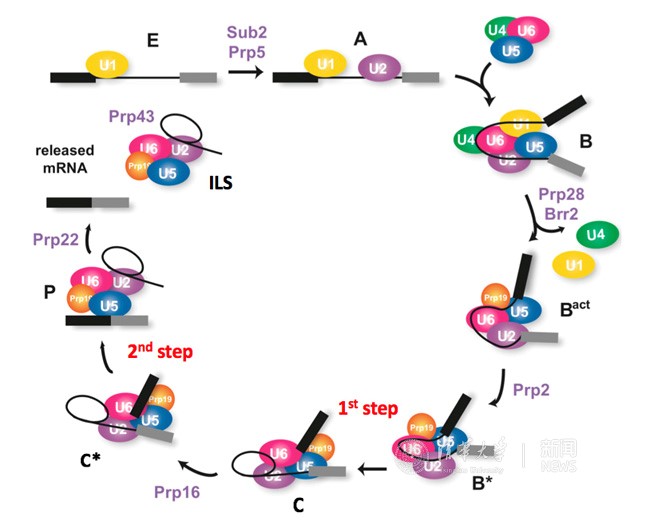
Figure 1. The splicing pathway. (modified from Janine, 2012)
Structural information is essential for understanding the mechanism of pre-mRNA splicing. In the summer of 2015, Professor Shi’s group reported the first atomic structure of a spliceosome from the yeast S. pombe, which represents the ILS state. In the following year, the Shi group reported a series of structures of spliceosomes from S. cerevisiae at distinct stages, including the activated Bact complex, the C complex, and the step II catalytically activated C* complex, as well as a major spliceosomal component, the U4/U6.U5 tri-snRNP. These high resolution structures have revealed unprecedented mechanistic insights into pre-mRNA splicing, corroborating and explaining a large body of genetic and biochemical data.
Despite the tremendous success on obtaining atomic three-dimensional structures of the yeast spliceosome, the human spliceosome has eluded structural determination at atomic resolution as a consequence of its inherent compositional heterogeneity and conformational flexibility. Considering the fact that more than 1/3 of human genetic diseases are directly correlated to splicing defects, the characterization of the human spliceosome structure will not only improve our understanding of the biochemical nature of pre-mRNA splicing but also illuminate the potential therapies for many hereditary diseases and further assist in structure based drug discovery targeting the spliceosome.
In the recently published Cell article, researchers adopted a modified pre-mRNA to assemble the human spliceosome in vitro and have successfully stalled the reaction at a specific stage, after the Step1 transesterification and prior to the exon ligation. The conformational flexibility of the human spliceosome was minimized by the utilization of chemical crosslinking (GRAFIX) under physiological conditions. The resultant pure and homogeneous sample enabled the structure determination of human spliceosome at 3.76 angstrom resolution. The resolution of the core of spliceosome, which consists of approximately 20 components, reaches to a resolution of 3.0-3.5 angstrom, allowing the unambiguous observation of the conformation of the active sites. Several step2 factors adopts conformations that either stabilize the active sites or promote the step2 catalytic reaction (Figure 2 and Figure 3). These structural features reveal important mechanistic insights into the conformational changes of the spliceosome and recognition of the 3’ splicing sites during the step2 catalytic reaction. Most notably, this is the first reported atomic structure of the human spliceosome.
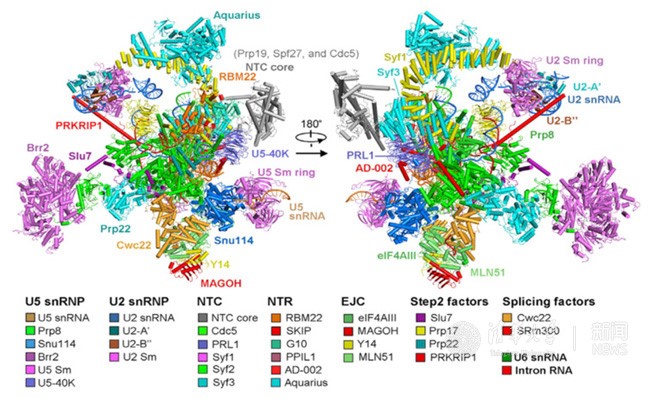
Figure 2. Structure of the human C* spliceosomal complex
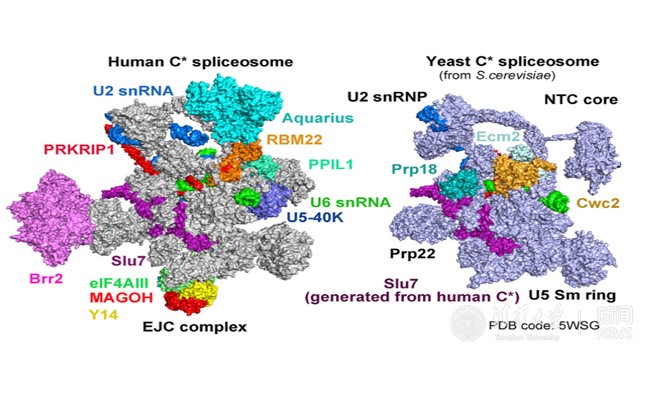
Figure 3. Comparison of the human and yeast C* spliceosomal complexes
Professor Yigong Shi is the corresponding author; Xiaofeng Zhang, Dr. Chuangye Yan and Jing Hang are the co-first authors; Dr. Chuangye Yan is also the co-corresponding author; Dr. Jianlin Lei, the director of Cryo-EM facility of Tsinghua University, provided many valuable advices and assistance on data collection; Dr. Xiaomin Li and Dr. Xiaomei Li also provide important assistance on data collection; Dr. Lorenzo I. Finci made initial contributions to this project; Lijia Li participated in this project. The project was supported by the cryo-EM facility of Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) and the computational facility on the cluster of the Bio- Computing Platform. This work was supported by funds from the National Natural Science Foundation of China and the Ministry of Science and Technology.
Website of this article:
http://www.cell.com/cell/fulltext/S0092-8674(17)30487-7
Structures of the yeast spliceosomes reported by the Shi group:
http://science.sciencemag.org/content/early/2016/12/14/science.aak9979.full
http://science.sciencemag.org/content/early/2016/01/06/science.aad6466
http://science.sciencemag.org/content/early/2015/08/19/science.aac8159
http://science.sciencemag.org/content/early/2015/08/19/science.aac7629
http://science.sciencemag.org/content/early/2016/07/20/science.aag0291
http://science.sciencemag.org/content/early/2016/07/20/science.aag2235

