Breakthrough in Ribosome Biogenesis: Atomic Structures of the Pre-Ribosome Revealed by Tsinghua cryo-EM team
Research group of Prof. Ning Gao at Tsinghua University and their collaborator Prof. John Woolford at Carnegie Mellon University recently published a research paper on Nature, entitled “Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes”.
Ribosome biogenesis in eukaryotes is an extremely complex process regulated by hundreds of trans-acting factors, including energy-consuming enzymes, RNA chaperones and RNA/protein modification enzymes. The regulation of ribosome assembly is tightly coupled with various cellular pathways governing the cell growth and proliferation. Mutations in some of the assembly factors lead to dysregulation of ribosome assembly and contribute to a diverse collection of human diseases named as ribosomopathies. However, the functions and structures of most of these assembly factors remain unclear.
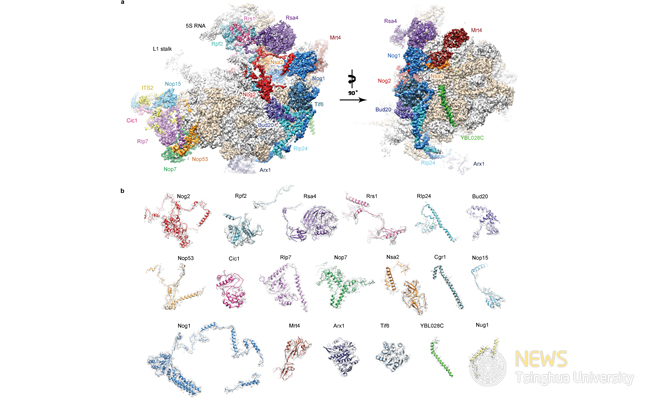
Figure 1 Cryo-EM structure (state 1) of the pre-60S particle purified from epitope-tagged Nog2. a, The 3.08- A cryo-EM map of state 1 is displayed in surface representation, with density of each assembly factor separately coloured. The 25S rRNA and r-proteins are coloured grey and beige, respectively. Both the intersubunit (left) and side (right) views are shown. b, Atomic models of 19 well-resolved assembly factors (coloured as in a) superimposed with their segmented cryo-EM densities (transparent grey).
In this paper, the authors used an assembly factor Nog2 as the bait to purify endogenous ribosome assembly intermediates from Saccharomyces cerevisiae, and employed the state-of-art cryo-EM and mass spectrometry techniques to characterize the structure and composition of the purified heterogeneous pre-60S ribosomal particles. Structures of the pre-60S ribosomes in different functional states were subsequently obtained. One specific state (State 1) was solved at a nominal resolution of 3.08 A and atomic models of 19 assembly factors were built (Fig. 1).
The rich structural information in their structures provides a framework to dissect molecular roles of diverse assembly factors in eukaryotic ribosome assembly. Particularly, the structural data on two essential GTPase Nog1 and Nog2 shed light on their critical roles in structural remodeling checkpoints and nuclear export of the pre-60S ribosomal particles.
Prof. Ning Gao and Prof. John L. Woolford Jr at Carnegie Mellon University are corresponding authors. Miss Shan Wu (2013 doctoral student in the school of Life Sciences, Tsinghua University) is the first author of paper. Prof. Meng-Qiu Dong and Dr. Dan Tan from National Institute of Biological Sciences, Beijing, provided CXMS data. Cryo-EM data collection and computation were supported by the National Center for Protein Sciences (Beijing, China). The work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China, Beijing Advanced Innovation Center for Structural Biology, and National Institute of Health (USA).
Article link: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature17942.html

