Prof. Jijie Chai’s group and Prof. Sen-Fang Sui’s group reported structural and biochemical investigations on the activation mechanism of NAIP-NLRC4 inflammasome in Science
On Oct 8th, the research teams led by Prof. Jijie Chai and Prof. Sen-Fang Sui in School of Life Sciences, Tsinghua University, published a research article in Science by earlier publication online, reporting the structure of a PrgJ-NAIP2-NLRC4 inflammasome at 6.6 angstrom resolution determined by single particle cryo-electron microscopy (cryo-EM), and revealing the induced self-propagated activation mechanism of NLRC4.
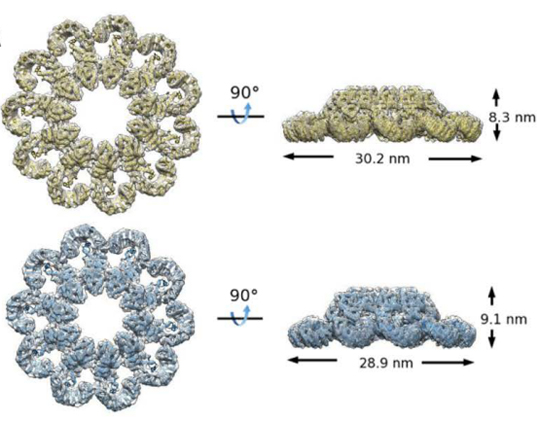
Figure 1. The three-dimensional structure of PrgJ-NAIP2-NLRC4ΔCARD complex
Nucleotide-binding domain (NBD)- and leucine-rich repeat (LRR)-containing proteins (NLRs) are critical for the cytosolic immunosurveillance system of mammals. Dysregulation of NLR function is associated with several human diseases. NLRs are pattern recognition receptors that recognize pathogen-associated molecular patterns (PAMPs) or host-derived danger components, resulting in NLR activation and oligomerization. The oligomerized NLRs then recruit procaspase-1 (pro-Casp1) either directly or through the adaptor protein apoptosis-associated speck-like protein containing CARD (caspase activation and recruitment domain) (ASC), forming inflammasomes that catalyze Casp-1 activation. Once activated, Casp-1 proteolytically processes pro-interleukin 1β (pro-IL-1β) and pro-IL-18 to initiate host innate immune responses.
NLRC4 (NOD-like receptor containing CARD 4), one member of NLRs, recognizes the cytosolic flagellin and the rod protein of type III secretion system. In the resting state, NLRC4 is autoinhibited with the mechanism being elucidated by our previous work (http://www.sciencemag.org/content/341/6142/172.full). Once sensing PAMPs, NLRC4 is activated and oligomerizes into multi-protein signaling complexes termed inflammasomes, activating the inflammation and other immune reactions. NLRC4 containing inflammasomes are activated by bacterial pathogens carrying flagellin or the components of type III secretion system (T3SS). Specificity for different bacterial ligands is dictated by NLR apoptosis inhibitory proteins (NAIPs). In mice, direct recognition of the bacterial flagellin and the T3SS rod protein PrgJ is mediated by NAIP5/6 and NAIP2, respectively. Ligand binding induces NAIP interaction with NLRC4 followed by oligomerization of their complex. However, the molecular mechanism underlying the activation of NAIP-NLRC4 inflammasome is poorly understood.
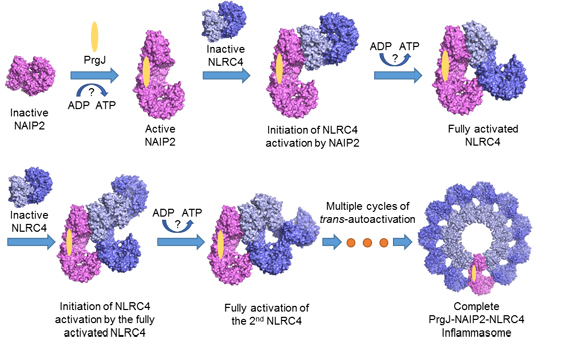
Figure 2. A schematic diagram for PrgJ-induced assembly of the wheel-like structure of a PrgJ-NAIP2-NLRC4 complex
In this study, the research teams led by Prof. Chai and Prof. Sui determined the wheel-like structure of PrgJ-NAIP2-NLRC4ΔCARD complex at 6.6 angstrom resolution using the most advanced cryo-EM reconstitution techniques (Figure 1). The activation of NLRC4 involves substantial structural reorganization that creates one oligomerization surface (catalytic surface). Once activated, NLRC4 utilizes this surface to catalyze the activation of an inactive NLRC4, self-propagating its active conformation to form the wheel-like architecture. NAIP proteins possess a catalytic surface matching the other oligomerization surface (receptor surface) of NLRC4 but not those of their own, ensuring that one NAIP is sufficient to initiate NLRC4 oligomerization.
Our structural analysis, together with the biochemical studies, support a sequential ligand-induced assembly of NAIP-NLRC4 inflammasomes (Figure 2). The properties of NLRC4 oligomerization are reminiscent of the replication mechanism of a prion in which conformational conversion from its properly folded isoform into the prion form (misfolded) is propagated in an autocatalytic manner. Such a mechanism would render hosts more efficient to sense the danger signals and initiate the immune responses.
Dr. Zehan Hu, Dr. Qiang Zhou and Chenlu Zhang (PhD student) from School of Life Sciences, Tsinghua University, are co-first authors of this paper. Prof. Jijie Chai and Prof. Sen-Fang Sui from School of Life Sciences, Tsinghua University, are corresponding authors of the paper.
This work was supported by funds from the Ministry of Science and Technology, the National Natural Science Foundation of China, Tsinghua-Peking Center for Life Sciences and China Postdoctoral Science Foundation. The EM data were acquired on the Tsinghua Cryo-EM Facility and processed on the Explorer 100 cluster system of Tsinghua National Laboratory for Information Science and Technology.
Web link of this paper:
www.sciencemag.org/content/early/2015/10/07/science.aac5489.full

