Prof. Yigong Shi’s group reported the structure of yeast spliceosome and the splicing mechanism in two Science articles
On August 21st, the research team led by Prof. Yigong SHI from School of Life Sciences, Tsinghua University published two side-by-side research articles in Science, reporting the long-sought-after structure of a yeast spliceosome at 3.6 angstrom resolution determined by single particle cryo-electron microscopy (cryo-EM), and the molecular mechanism of pre-messenger RNA splicing.
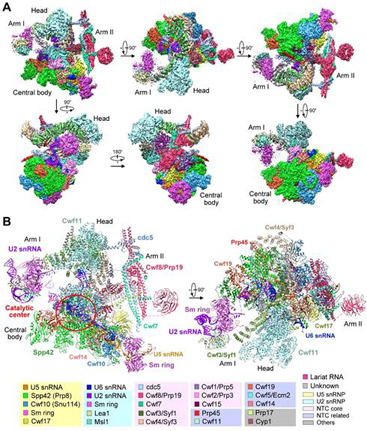
Figure 1 : The three-dimensional structure of a yeast spliceosome at 3.6 angstrom resolution.
Gene expression is the basic principle of all living cells. During the process known as the Central Dogma, the genomic information stored in genome DNA sequences is delivered to pre-mRNAs by transcription and finally to functional proteins by translation. In eukaryotes, pre-mRNAs are intervened by coding sequences containing exons and untranslated introns. The excision of introns and ligation of exons is named as pre-mRNA splicing, and executed by spliceosome. This macromolecular machinery consists of five small nuclear ribonucleoprotein particles (snRNPs), nineteen complex (NTC), NTC related (NTR), and a number of associated enzymes and cofactors. In total, more than 100 proteins and at least 5 RNAs were identified to be the core and auxiliary components of spliceosome. The involved proteins and RNAs assemble into and dissociate from spliceosome in a strict order during splicing, endowing extreme dynamics and flexibility of the spliceosome. These features guarantee the accomplishment of the complex splicing reaction, but at the same time tangling the structural investigations of spliceosome.
Besides the basic biological importance of spliceosome, numerous diseases are related to the dysfunction of spliceosomal regulation or the splicing mistakes. Almost 35 percent of genetic disorder is resulted from wrong splicing, exemplified by unusual expression of alternative splicing that leads to frontotemporal dementia driven by tau mis-splicing. The mutation of key spliceosomal proteins like Brr2 or Prp8 can lead to the Autosomal Dominant retinitis pigmentosa. Some cancer is also associated with abnormal splicing.
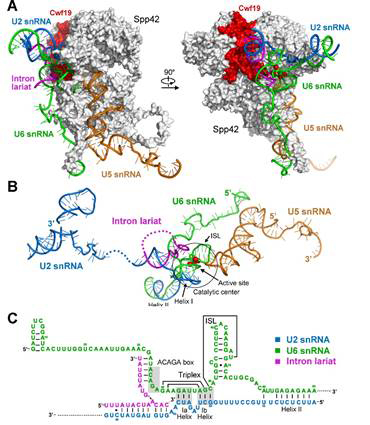
Figure 2: The diagram for molecular basis of pre-mRNA splicing.
Prof. SHI has long been intrigued by the mechanism of splicing and his group has been working on the spliceosome way back in 2009. In 2014, SHI’s group reported in NATURE the first crystal structure of the Lsm complex, which is a key component of the spliceosome. Driven by his ambition and not ceased by the immediate success, Prof SHI’s group continued the research on the intact spliceosome. This year, they have reached a major milestone in obtaining the endogenous intact yeast spliceosome with high purity. This was made possible by the substantial improvement on the tandem-affinity purification. Using the most advanced cryo-EM reconstitution techniques, the spectacular 3-D atomic model was resolved, which consists of 10,574 amino acids from 37 proteins and 4 RNA molecules. The combined molecular mass was approximately 1.3 mega-Daltons. Based on the structural analysis together with the existing knowledge, the group provided the first structural insight into the molecular mechanism for pre-mRNA splicing: the spliceosome is in essence a protein-directed ribozyme, with the protein components essential for the delivery of critical RNA molecules into close proximity of one another at the right time for the splicing reaction (shown in Figure 2).
Since the discovery of “split genes” in 1977, scientists are constantly exploring the molecular mechanism of pre-mRNA splicing. In 1983, Steitz, J.A. group isolated five U snRNPs; in the same year, Sharp, P.A. and Keller, W group set up the in vitro splicing assay independently. Until now, decades of genetic and biochemical experiments have identified almost all proteins in spliceosome and uncovered some functions. Yet, the structure remained a mystery for a long time. The works, primarily performed by Dr. Chuangye Yan, and Ph.D students Jing Hang and Ruixue Wan under Prof. Yigong Shi’s supervision, settled this Holy Grail question and established the structural basis for the related area.
This work was supported by funds from the Ministry of Science and Technology and the National Natural Science Foundation of China. The EM data were acquired on the Tsinghua Cryo-EM Facility and processed on the Explorer 100 cluster system of Tsinghua National Laboratory for Information Science and Technology.

