Yigong Shi’s group published the structure of human g-secretase at atomic resolution
Prof. Yigong Shi’s laboratory in School of Life Sciences, Tsinghua University, reported the 3.4 angstrom cryo-EM structure of γ-secretase in a research article entitled “An atomic resolution structure of human γ-secretase” published online in Nature on August 17th 2015. This work, in collaboration with Drs. Xiaochen Bai and Sjors Scheres from the Laboratory of Molecular Biology of MRC, UK, provides an important framework for understanding the disease mechanism of γ-secretase and Alzheimer’s disease.
Dysfunction of the intramembrane protease γ-secretase is thought to cause Alzheimer’s disease (AD), with most mutations derived from AD mapping to the catalytic subunit presenilin 1 (PS1). Prof. Shi and co-workers reported an atomic structure of human γ-secretase at 3.4 angstrom resolution, determined by single-particle cryo-electron microscopy. Mutations derived from Alzheimer’s disease affect residues at two hotspots in PS1, each located at the centre of a distinct four transmembrane segment (TM) bundle. TM2 and, to a lesser extent, TM6 exhibit considerable flexibility, yielding a plastic active site and adaptable surrounding elements. The active site of PS1 is accessible from the convex side of the TM horseshoe, suggesting considerable conformational changes in nicastrin extracellular domain after substrate recruitment. Component protein APH-1 serves as a scaffold, anchoring the lone transmembrane helix from nicastrin and supporting the flexible conformation of PS1. Ordered phospholipids stabilize the complex inside the membrane. This structure serves as a molecular basis for mechanistic understanding of γ-secretase function.
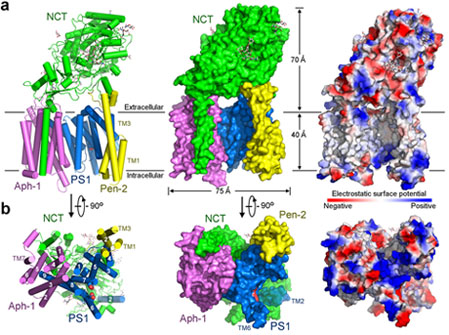
(Figure 1. The atomic structure of human g-secretase.)
It is noteworthy that structural characterization of γ-secretase has been very slow due to the daunting technical challenges concerning expression and purification of the intact γ-secretase complex, which was considered to have 19 transmembrane helices. The Shi Lab has launched rigorous and systematic efforts for structural characterization of γ-secretase and mechanistic understanding of AD in the past decade. Obtaining homogeneous and active human γ-secretase for structural investigation proved to be the bottleneck. After numerous trials and tribulations, they eventually made great breakthrough in recent years.
December, 2012: the Shi Lab reported the crystal structure of PSH, a bacterial homologue of PS1, which revealed a novel fold. A homologous structural model of PS1 was generated and the AD-related mutations were for the first time mapped to the structural model (Li et al, Nature, 2012).
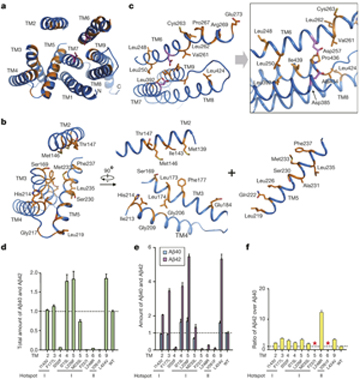
(Figure 2. Alzheimer’s disease-derived mutations mapped to two hotspots on PS1.)
June, 2014: the Shi Lab reported the 4.5- angstrom cryo-EM structure of the human γ-secretase in collaboration with Drs. Xiaochen Bai and Sjors Scheres from the Laboratory of Molecular Biology, Molecular Research Council at Cambridge, UK. The transmembrane helices can be distinguished, but assignment of each subunit awaited further improved resolution (Lu et al, Nature, 2014).
September, 2014: the Shi Lab reported the 1.95- angstrom crystal structure of the extracellular domain (ECD) of a Nicastrin homolog. Nicastrin is responsible for substrate recruitment for γ-secretase. Based on this structure, they were able to build the homologous model of human Nicastrin ECD, which facilitated domain assignment of the 20 transmembrane helices observed in the previous EM map (Xie et al, PNAS, 2014).
March, 2015: the Shi lab reported the biochemical characterization of PSH, which suggested that PSH shows similar cleavage activity with γ-secretase and is subject to regulation by γ-secretase inhibitors. They also reported the structure of PSH in complex with inhibitor. This structure shows the binding pocket of this inhibitor thus allowing the preliminary screen of γ-secretase inhibitors and drugs with less investment in harvesting the protein (Dang et al, PNAS, 2015).
April, 2015: the Shi lab reported the cryo-EM structure of human γ-secretase at 4.3 angstrom resolution. Despite the slightly improved overall resolution, the density in the transmembrane region was considerably better resolved. More importantly, fusion of the N-terminus of PS1 with a T4 lysozyme led to unambiguous identification of the first transmembrane helix of PS1 and subsequent assignment of all the four components of γ-secretase in the transmembrane region. It confirmed the previous subunit assignment. In addition, the protein sample was prepared in a mild detergent digitonin. The similar structure with the previous one determined in amphipol alleviated the concern that the structure may be distorted by amphipol or sample preparation (Sun et al, PNAS, 2015).
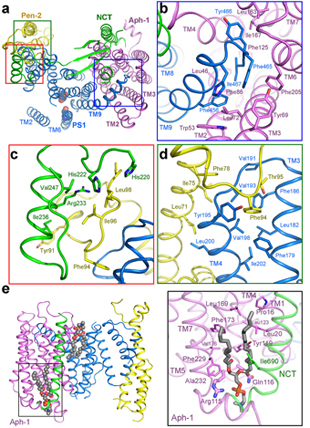
(Figure 3. Assembly interfaces among the four components of g-secretase in the transmembrane region)
The most recent report on the structure of γ-secretase at 3.4 angstrom resolution was accomplished from the continued collaboration between the Shi Lab and the MRC group. The detailed structure provides an important framework for the study of function and disease mechanism of γ-secretase as well as Alzheimer’s disease.
Dr. Xiaochen Bai, who has been a postdoctoral fellow in LMB of MRC after his PhD study in Tsinghua University, Dr. Chuangye Yan, and Guanghui Yang a 3rd year PhD candidate in School of Life Sciences, Tsinghua University, are co-first authors.
This work was supported by funds from the Ministry of Science and Technology (2014ZX09507003006 to Y.S.), the National Natural Science Foundation of China (31130002 and 31321062 to Y.S.), Centre for Life Science, a European Union Marie Curie Fellowship (to X.C.B.), and the UK Medical Research Council (MC_UP_A025_1013, to S.H.W.S.).
The paper links:
http://www.nature.com/nature/journal/v493/n7430/abs/nature11801.html
http://www.nature.com/nature/journal/v512/n7513/full/nature13567.html
http://www.pnas.org/content/111/37/13349.short
http://www.pnas.org/content/112/11/3344.short
http://www.pnas.org/content/112/19/6003.long
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature14892.html

